Cannalingo Part II: Connecting Molecular Conversations of CB2R with the Cannabis Experience
Part II of a guided tour through the protein structure and function influence of cannabinoid receptor 2 (CB2R) on cannabinoid signaling.
Our journey through the molecular maze of cannabinoid receptor signaling continues with an in-depth exploration of the structure, function, and variants of cannabinoid receptor 2 (CB2R). The second of two characterized cannabinoid receptors, CB2R, plays a significant, albeit lesser, role in cannabinoid signaling compared to CB1R. Of the five variants of CB2R that have been characterized in humans, two are silent, meaning that there is no change to the protein. Three of the variants are missense mutations, resulting in changes to the protein sequence, and potentially function. It is hypothesized that slight changes in the chemistry of these three amino acids may impact the cytoplasmic function and the internal signaling of the protein. The impact may influence the functional response to cannabinoids for an individual who expresses a CB2R variant. The structural and functional impact of these three variants will be considered as the role of CB2R in cannabinoid signaling is explored.
CBR2 was the second cannabinoid binding receptor identified in humans. It is a 360 amino acid protein that shares 44% of the same amino acid sequence as CB1R. Like CB1R, CB2R is also expressed in the cell membrane, where it awaits binding to ligands and becomes activated to initiate a series of signals that ultimately induces the effects of cannabinoids both foreign and endogenously synthesized. CB2R is more conserved across species, generally meaning the sequence of this protein has not changed or mutated much since the species who express this protein parted ways on the evolutionary tree.
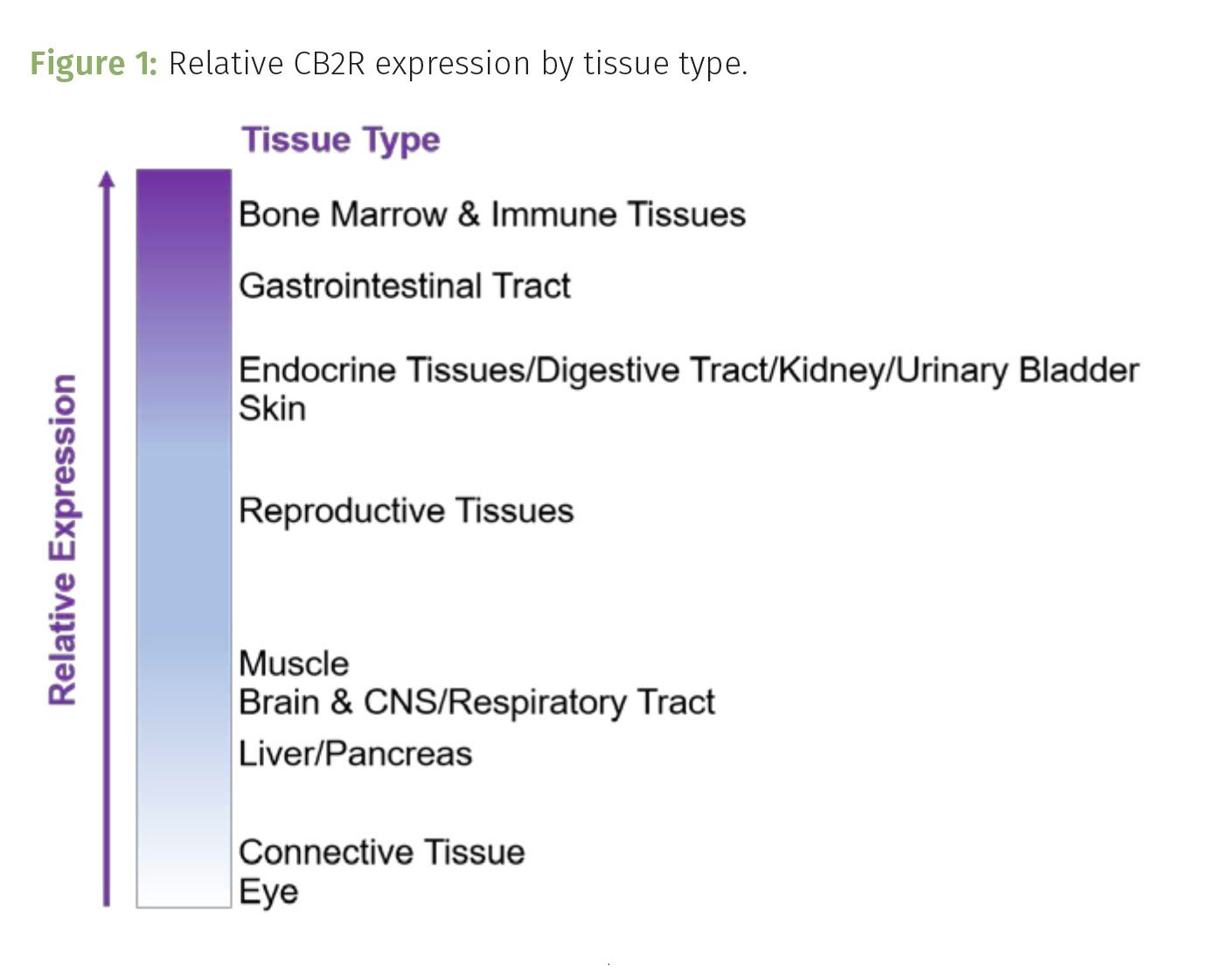
We can find CB2R expressed in neuronal, glial, and endothelial cells in the brain where it contributes to the regulation of neuronal activity (1). It is most abundant in immune tissue such as the spleen, tonsils and thymus, cardiovascular and respiratory system, and reproductive tissue including the testis (Figure 1) (2,3). While CB2R is not expressed as abundantly as CB1R, the expression of CB2R is inducible, meaning that certain stimulation can cause this receptor to increase its expression within the membrane. While this might sound exciting, this type of stimulation is not usually like the ones we seek out for fun. Addiction, inflammation, anxiety, and epilepsy serve to stimulate CB2R expression (4). This explains why CB2R is such a popular target of emerging drug discovery efforts. CB2R serves as a possible disease or condition-associated target for potential therapeutic treatments to prevent inducing its expression with over stimulation caused by conditions such as anxiety and epilepsy.
The endocannabinoid 2-arachidonoylglycerol (2-AG) binds to and activates CB2R, while anandamide (AEA) does not. What’s interesting is, 2-AG basal expression is 1000x higher than AEA in the brain. This abundance in expression contributes to the inducible nature of CB2R. Delta-9-tetrahydrocannabinol (∆9-THC) binds readily to CB2R, while cannabidiol (CBD) does not. Signaling by CB2R is impacted by THC products, rather than CBD products.
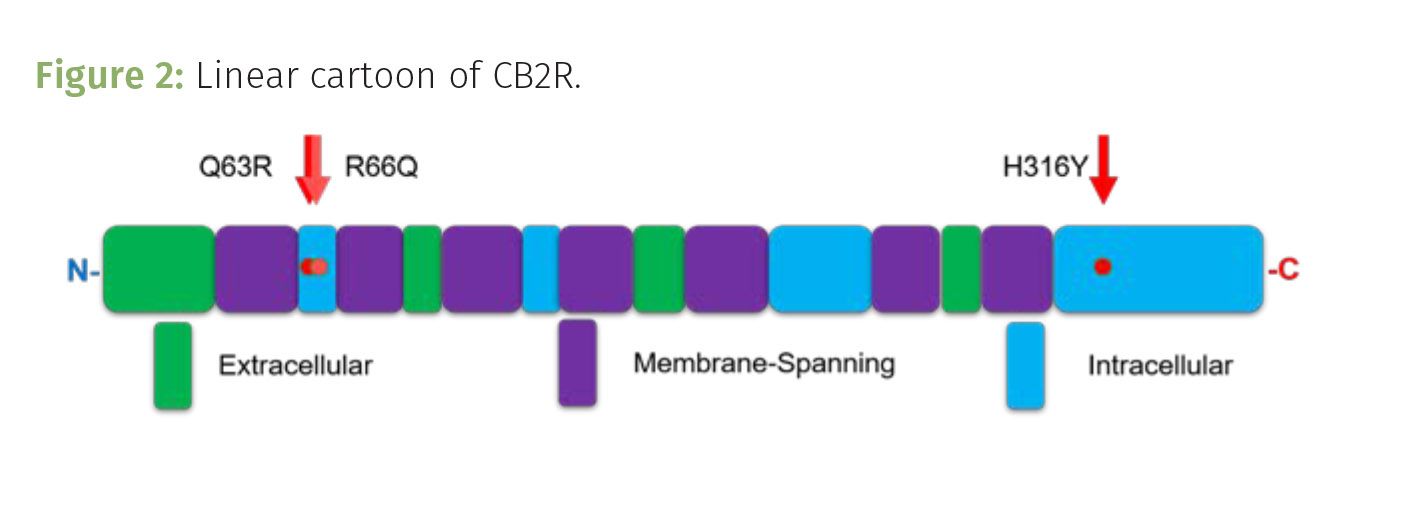
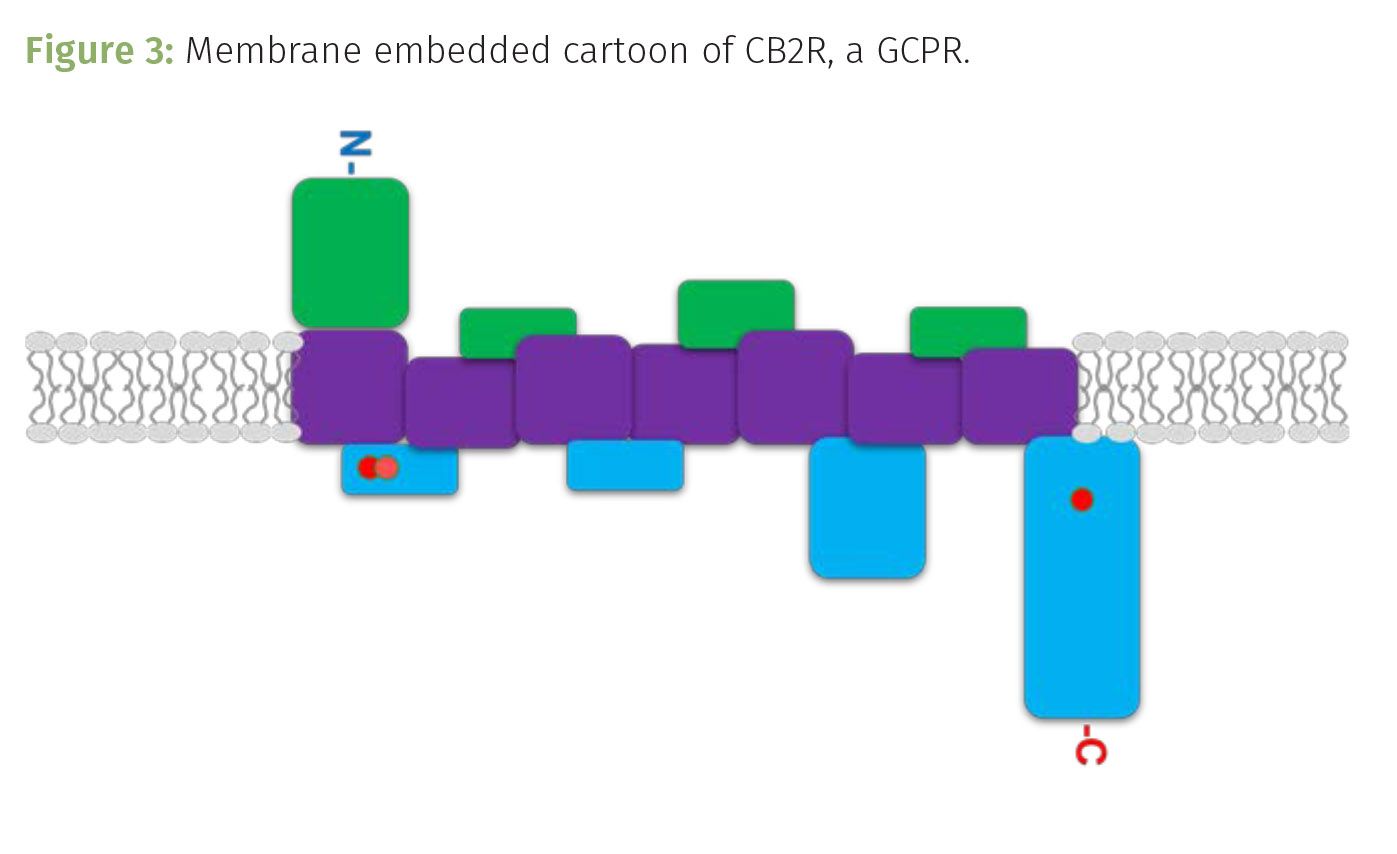
CB2R is encoded by the CNR2 gene located on chromosome 1. When expressed, it serves as a membrane embedded, G-coupled protein receptor (GCPR). Once activated by binding to a ligand, such as THC, the intracellular components of the receptor move to seemingly kick an intracellular G-protein into activation. This results in a domino effect, amplifying cell signaling that changes the cell’s behavior thereby inducing the analgesic and other effects of cannabinoid signaling. CB2R’s structure is typical of G-coupled protein receptors, including seven transmembrane domains that serpentine through the cell membrane, with four extracellular domains and four cytoplasmic, intracellular loops (Figure 2). The extracellular domains interact with ligands outside of the cell, such as THC, while the intracellular loops communicate the cellular response from the receptor being activated by the ligand binding to it (Figure 3).
There are two versions of CB2R expressed in humans: CB2A and CB2B. CB2A is a slightly longer version of the two, having a longer sequence at the start of the receptor. CB2A is predominantly expressed in the testis and to a lesser extent other tissues including the central nervous system and immune system. Whereas CB2B is predominantly expressed in the spleen and immune cells, followed by other peripheral tissues. How the CB2A structure and function changes because of the increase in length is yet to be determined. Some evidence suggests that the longer isoform may demonstrate more sensitivity to increased expression when induced by receptor agonists.
Similar to chromosome 6 in the CB1R story, chromosome 1 has also been demonstrated to have duplication events; extra copies of gene sequences may be present on the chromosome of certain individuals. However, these duplication events have not been observed in the portion of the chromosome that encodes for CB2R. Rather, the variant story associated with CB2R is far more exciting. Five variants have been characterized among people. To appreciate the intricacies of these variants, a short explanation about proteins is in store.
Proteins, the working molecules of life, are polymers assembled by building blocks of amino acids. There are 20 different amino acids found in proteins that support life. These amino acids differ in their chemistry; some are polar (hydrophilic, water-loving) and readily interact with the aqueous external and internal cellular environment. Hydrophilic dominant protein sequences will take on a three-dimensional (3D) shape that maximizes their interaction with water and other polar molecules. Other amino acids are nonpolar (hydrophobic or water-fearing), and exclude themselves from aqueous environments; hydrophobic dominating sequences of proteins often found in lipid filled membranes. Then, there are amino acids that can acquire a charge at physiological and more extreme pH. These amino acids can be negative or positively charged and often confer support for building the 3D shape or interacting with other molecules through ionic binding (the formation of salt bridges).
Proteins evolved to perform specific reactions to support the biochemistry of life. While proteins can be relatively short—a few 100 amino acids—to incredibly large—tens of thousands of amino acids—not all possible sequences of amino acids are observed in life. The energetic stability, as well as the ability of the resulting structure to support the given function of the protein dictate whether a protein is expressed and utilized to support life. Since life first began as single-celled organisms, life has been bombarded with exposures that mutate or change the DNA sequence that encodes for proteins. Some of the mutations resulted in variants of the protein that acquired a more efficient function, while others were removed from the population because the mutation did not support life. Over time, the mutations accumulated, duplication or removal of genetic material occurred, which ultimately lead to the production of new genes and proteins as the amino acid sequence changed, substituting, inserting, duplicating, or deleting amino acids along the way. As these new working molecules were introduced, new functions and interactions contributed to more sophisticated cellular functions and life forms. Fortunately, the atmosphere on Earth has become more hospitable since life first evolved and we have acquired some molecular machinery that can help life overcome certain mutations. However, we still find variation among the population, imposed by natural errors in cellular replication or induced by exposures to things such as ultraviolet (UV) light damage or carcinogens. Regardless, protein variants persist.
Two of the CB2R variants identified, F97F and C313C are silent mutations. Meaning that while the DNA sequence of the gene has changed, the amino acid encoded for by that part of the gene has remained the same. Position 97 of the amino acid sequence is still phenylalanine and 313 is still cysteine. There is no change in the amino acid sequence or shape of the protein, but this also goes to show that not all mutations are bad.
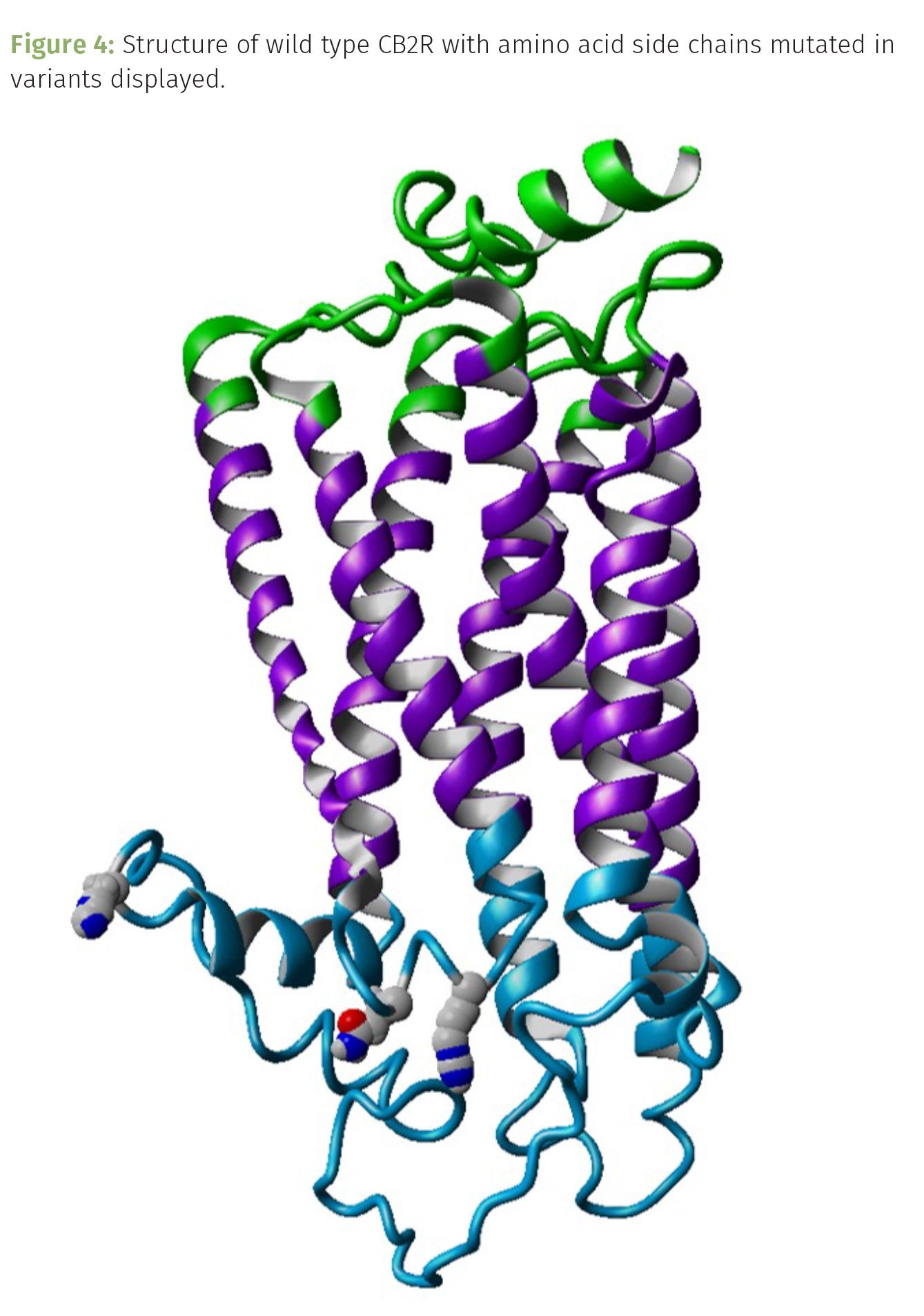
The other three variants—Q63R, R66Q, and H316Y—are a bit more interesting (Figure 4). These are missense mutations because the amino acid in the protein sequence changes, but these have the added intrigue of changing the chemistry of that position along the protein too. We can begin to explore the impact variants may have on CB2R structure and function by running molecular dynamic simulations (MDS). In other words, running modeling calculations to determine the changes in stability of the overall protein, and even pinpoint where in the sequence the structure may change. These are simulations, predictions of what may be occurring under ideal physiological conditions. While not grounded truth, MDS provide us with a preview of what may be. To run an MDS, we start with the amino acid sequence of the protein under study. Working with a protein whose structure has been determined always provides more probability in your comparisons. Fortunately, the structure for CB2R has been resolved and published. Next, we run some threading-based programs to predict the structure of the variants associated with CB2R. After scoring the possible structures, we can begin the MDS process. I did this on CB2R and these three variants, and found some compelling evidence to justify subsequent investigations into learning more about just how these variants contribute to the multiverse of CB2R signaling among people.
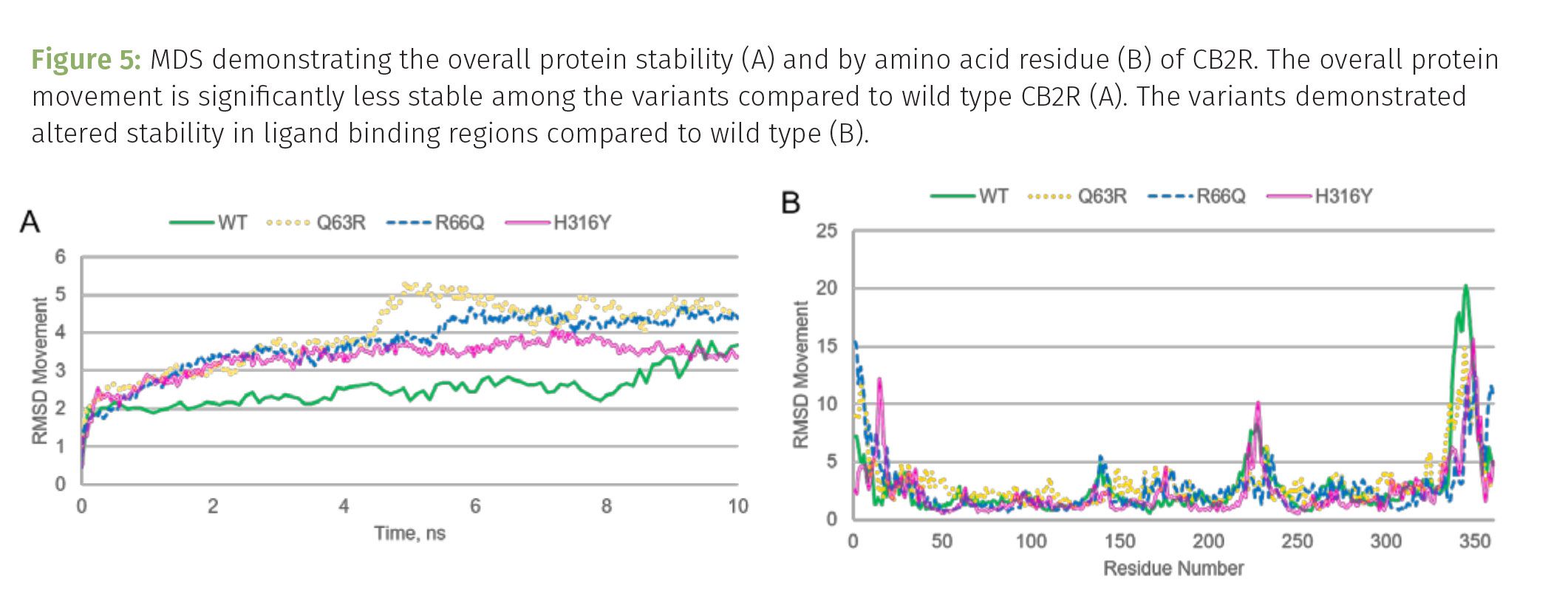
While all three variants are tolerable, an individual can survive these mutations, all three are significantly less stable than the wild type (original) protein (Figure 5A). The more movement observed as the protein takes on its shape during the first 10 ns of the simulation, the less energetically stable the protein. The first two variants at positions 63 and 66 of the sequence are both located in the first intracellular loop, while the variant at 316 is in the last intracellular portion of the protein (Figures 2 and 3). The first, Q63R, shorthand for glutamine, a polar amino acid, at position 63 is mutated to an arginine, a basic amino acid that is usually positively charged at physiological pH. This position of the protein is not super conserved among species, however the amino acid that is usually found here is either glutamine or arginine. This may seem like no big deal, however some hot off the press findings suggest that this variant may impact the severity of COVID-19, impacting the regulatory activity of endocannabinoids in immune cells, increasing the risk of inflammation. The Q63R variant may disrupt the G-protein’s ability to dock and be activated by the receptor (5). This does not unlock mysteries about why one individual may be more sensitive to cannabis exposure, but it sure does open the door for more research into the role this variant may play during our current pandemic life. The impact on structure is not incredibly extreme, especially right where the mutation is located. However, this change in chemistry may impact interactions the protein has with ligands in the extracellular domain (Figure 6B). More research is needed to sift through the impact of the variant further.
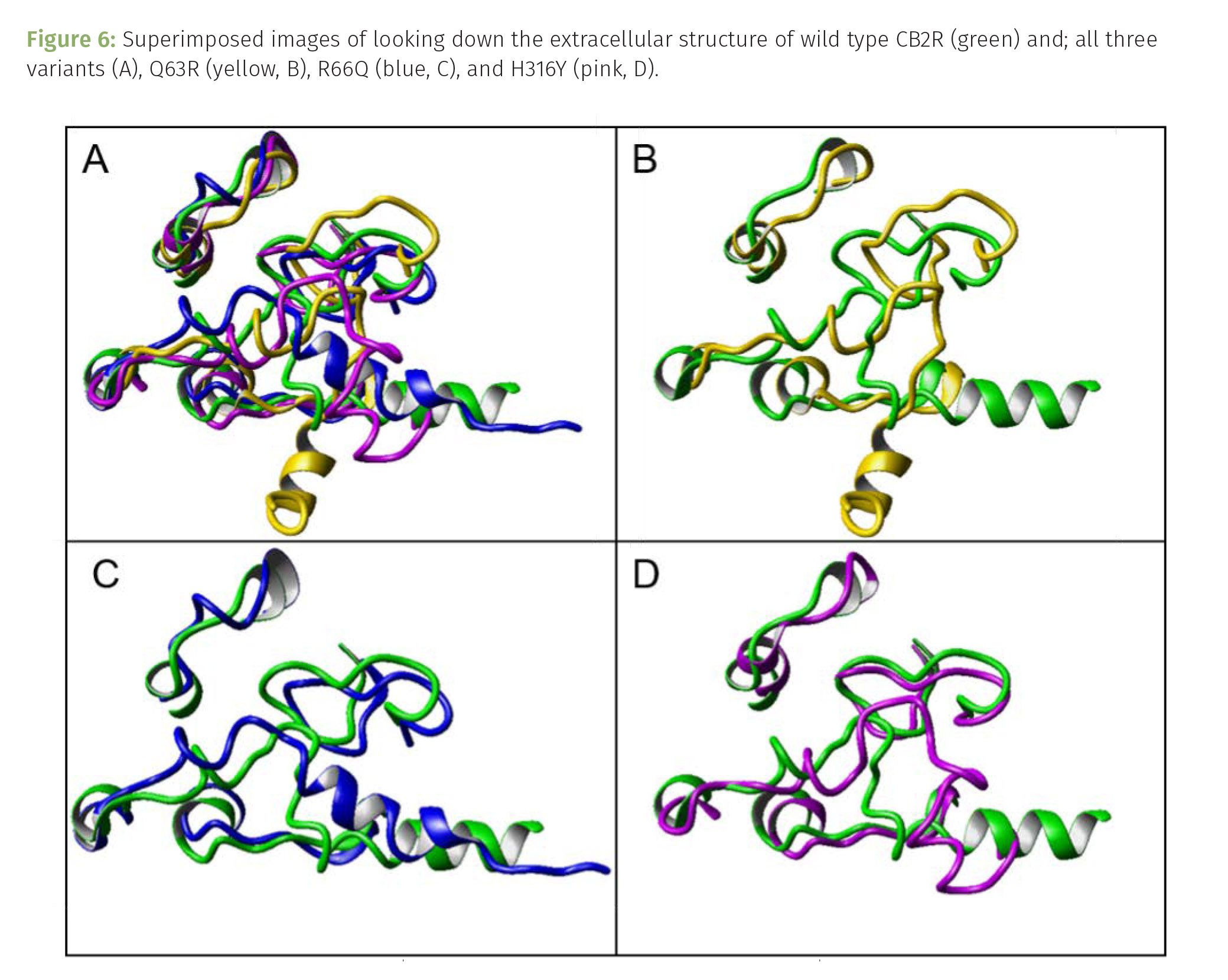
The R66Q variant is the opposite: an arginine at position 66 is mutated to a glutamine. Similarly, R66Q does not drastically change the structure of the protein at this intracellular loop, however the simulation suggests that ligand binding may be altered as well (Figure 6C). Unlike position 63, 66 is highly conserved across species. When it is not, the options for this position are generally arginine or glutamine. In fact, this arginine is part of a highly conserved sequence in CB2R, RSSVT (arginine, serine, serine, valine, threonine) that serves as a docking site for 14-3-3. When CB2R is bound to ligand and activated, 14-3-3 will bind to this region. This interaction may induce delays in the G2 to M cell cycle progression and slow down cellular replication. Disrupting this interaction could result in problems for the cell and subsequently derived cells. The progression from G2 to M serves as a checkpoint for ensuring that the cell is ready for mitosis (or meiosis depending upon the cell type). The cell must have enough energy, materials, and cellular machinery ready to support the process of dividing the cell into two daughter cells. Rushing through this phase may result in problems in the resulting daughter cells, which may be perpetuated in subsequent progeny from this faulty cell. Replacing the arginine in the RSSVT sequence may prevent the serines and threonine from becoming modified by phosphorylation. Enzymes target specific amino acids that can be phosphorylated (namely serine, threonine and tyrosines), adding a phospho group to an amino acid drastically changes its chemistry and often contributes to the interactions between other proteins, opening the docking site to support these interactions. The shape of the RSSVT sequence must complement the active site of the enzyme that adds the phospho group to the amino acid. The positive charge on arginine is significant to maintaining the complementary interactions with the enzyme. If the positively charged arginine is not in the sequence, QSSVT instead, the enzyme will not phosphorylate the serines or threonines as efficiently, 14-3-3 may not dock, and the cell can transition to M phase without conducting the appropriate quality control checks to make sure it is ready to do so. The impact this has on the cannabis experience is not quite elucidated yet, however other health consequences might be considered before teasing out this role.
The H316Y variant is characterized as a cyclic, polar, sometimes positive at fluctuations of pH common to the biochemistry of life amino acid to a cyclic, polar, and often phosphorylated amino acid. Histidine (H) and tyrosine (Y) share some characteristics yet are quite different amino acids. Different enzymes recognize, and physiological conditions influence, the phosphorylation of histidine versus tyrosine. This position along CB2R is found in the final intracellular domain and is significant in kickstarting the activation of the G-protein that initiates the resulting cellular signaling upon receptor activation. Activating the G-protein is an important part of this signaling pathway, so it is not surprising to learn that position 316 is in a highly conserved region of the protein. Fortunately, this variant appears to be well tolerated and must not impact the initiation of pathway too much. The MDS suggests that while the intracellular domain may not alter its shape too drastically, several potential ligand binding sites located on the extracellular domains of the receptor may be positioned differently (Figure 6D). Altering the shape of the ligand binding region of the receptor may impact cannabinoid binding efficiencies.
These are viable variants of CB2R. Albeit, there is some evidence to suggest that the 63 and 316 variants have been associated with autoimmune disorders. Laboratory-based studies of the Q63R and H316Y variants expressed in cell lines suggest that the variants can bind to cannabinoids and induce signal transduction. However, cannabinoid agonists, such as AG-2 and WIN55212-2 (a synthetic agonist), had reduced efficacy on the variants. Further, these CB2R variants demonstrated greater constitutive activity compared to the wild type receptor (6). All three variants share potential modifications to the ligand binding regions of the extracellular domains of the receptor (Figures 5B and 6). Superimposing the structures suggest that ligand binding ability of the variants may be altered, with H316Y having the most significant changes in shape (notice the alpha helix to the lower right of the wild type has become disordered in the H316Y variant).
Interest in studying the role CB2R and its variants has increased since expression of this receptor has been observed in brain and CNS tissues. Evidence suggests that CB2R may play a role in neuropsychiatric disorders such as depression, schizophrenia, and substance abuse. Clearly, more research is needed to better understand how variants of this receptor impact an individual’s response to cannabinoids.
References
- E.S. Onaivi, H. Ishiguro, S. Gu, and Q.R. Liu, J Psychopharmacol.26(1), 92-103 (2012).
- Q.R. Liu, C.H. Pan, A, Hishimoto, and et al., Genes Brain Behav. 8(5), 519-530 (2009).
- E.S. Onaivi, H. Ishiguro, J.P. Gong, and et al., Ann N Y Acad Sci. 1074, 514-536 (2006).
- J. Wu, Acta Pharmacol Sin. 40(3), 297-299 (2019).
- M. Rastegar, S. Samadizadeh, M. Yasaghi, and et al., Arch Virol. 166(11), 3117-3126 (2021).
- A. Carrasquer, N.M. Nebane, W.M. Williams, and Z.H. Song, Pharmacogenet Genomics. 20(3), 157-166 (2010).
About the Author
Audrey Shor is the co-founder and chief science officer of Decarb Factor in Denver, Colorado. Direct correspondence to: audrey@decarbfactor.com.
How to Cite this Article:
A. Shor, Cannabis Science and Technology® Vol. 5(5), 38-45 (2022).
